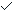For single file to single file conversion, you usually use mri_convert of freesurfer. For examplemri_convert x.img y.nii
Other options would be LONI Debabeler or MRICro.
Here are some special cases:
multiple 3D ANALYZE to a single 4D Nifti: mri_concat
mri_concat is a program of freesurfer. Assume the original image files are f001.img, f002.img … etc. The output file is f.nii.
-
mri_concat f???.img --o f.nii
mri_concat f???.img --o f.nii
You can also use FSL’s fslmerge to achieve the same thing (but pay attention to your fsl environment setting for default output format).
multiple DICOM to other format
To convert DICOM files to other format, use mri_convert
-
mri_convert I0001.dcm T1.mgz
mri_convert I0001.dcm T1.mgz
If you have a bunch of DICOM files (slices) for a single brain, you only need to input the 1st one as the argument of mri_convert.
Another way is to use SPM’s DICOM import. In the end you get ANALYZE files.
GE 7 file to ANALYZE or Nifti: makevols and makenifti
.7 files are functional images produced by GE scanners in Stanford. You use makevols to convert.
-
makevols E*P*.7 I
-
rename I.V I_ I.V*
makevols E*P*.7 I rename I.V I_ I.V*
You will get a bunch of .img/.hdr files.
To make a Nifti file, you run
-
makenifti Efilename outfilename
makenifti Efilename outfilename
The output is a single nii file (outfilename.nii).
Note: If you rename the .7 files, you might not get makevols to work. For example, if you rename P06144.7 to P06144_mer1.7, makevols doesn’t work (only produces a lot .hdr files but not .img files).
makevols and makenifti are written by Dr. Gary Glover at Stanford University. Here you can download the two programs.
Check out here to see how to convert images with different number of bytes per voxel.